News
November 9, 2025
The American Peptide Society highlighted our recent research in Nature Communications.
https://americanpeptidesociety.org/aps-news/peptide-portals/
August 6, 2025
Our story on a new antimicrobial peptide class is now available online! Thanks to all co-authors for a wonderful collaborative effort.
July 2, 2025
Congratulations to Fauzia for successfully completing her qualifying exams and advancing to candidacy.
March 4, 2025
Congratulations to Ama who advanced to the status of Doctoral Candidate after successfully passing her qualifying exam!
November 18, 2024
It's World AMR Awareness Week! Check out the research that we and our colleagues in AGNR are doing.
September 27, 2024
Congratulations to Dr. Bhowmick for her three minute flash talk presentation at the 2024 USM Postdoctoral Research Symposium!
June 7, 2024
We received our first NIH grant! An R21 to study the biosynthetic mechanisms for a new class of AMPs constructed at the cell membrane supported by the National Institute Of Allergy And Infectious Diseases.
December 18, 2023
Thank you AGNR for awarding a MAES research grant to our lab for the project, “Mechanistic analysis of a novel antimicrobial pore-forming peptide” . We hope to use this seed money to obtain external support for continuing our research!
Research
Methicillin-resistant Staphylococcus aureus (MRSA) represents a significant One-Health challenge, capable of spreading between and infecting both animals and humans. Our lab's research is focuses on understanding the physiology of MRSA to reveal potential targets for new drug development.
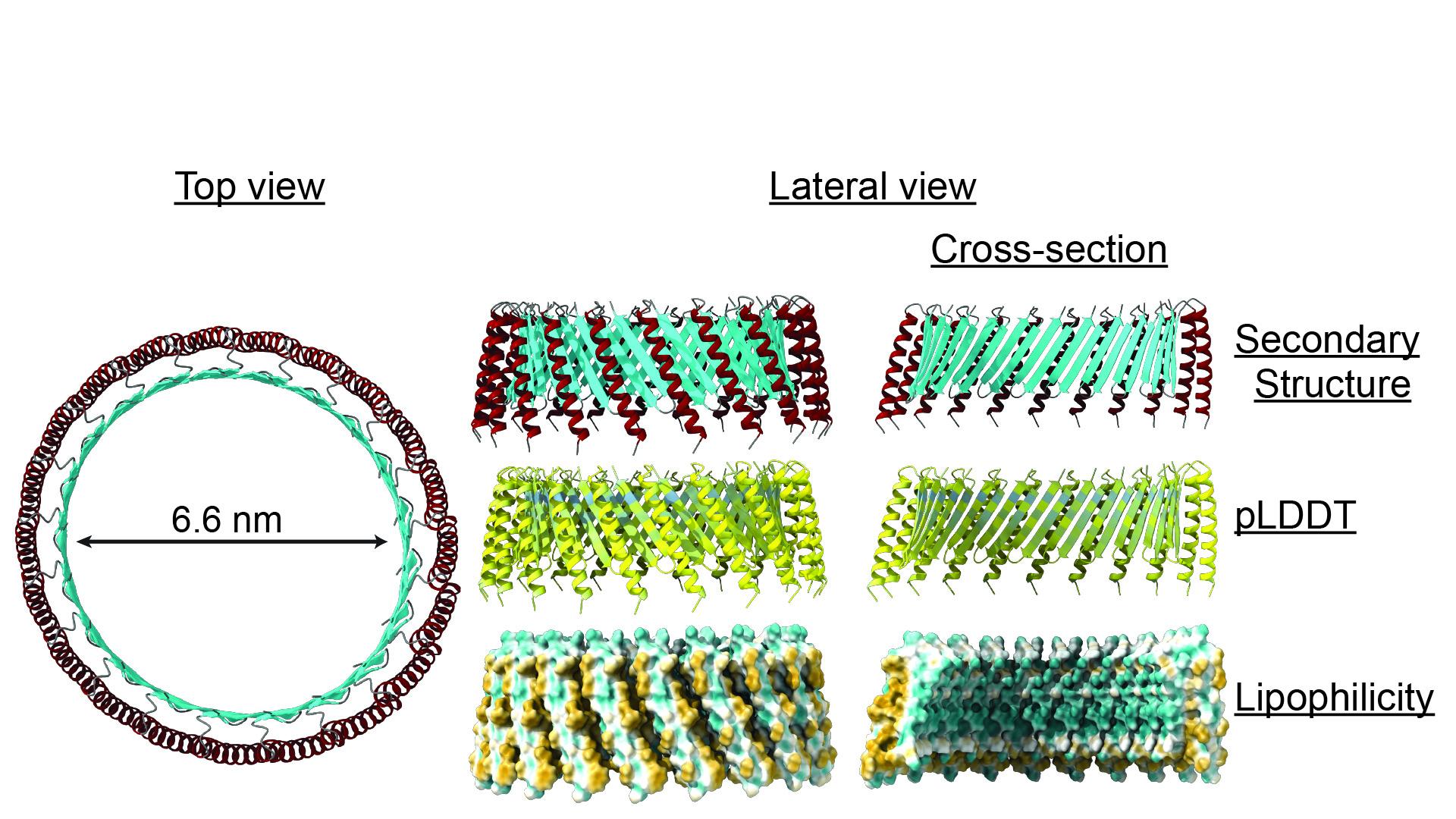
We are also exploring TMcin, a newly discovered pore-forming antimicrobial peptide (AMP) class that is active against MRSA and other Gram-positive pathogens. TMcin is unique among AMPs: it possesses a transmembrane-helix along with a two-stranded beta-sheet that allows it to reach target bacteria and form a large, oligomeric pore. The beta-sheets of oligomeric TMcins form a beta-barrel that spans the membrane and the transmembrane helices surround the barrel to anchor the pores in the cell membrane.
Antimicrobial Peptide Biosynthesis
How do cells secrete an antimicrobial peptide with a transmembrane helix? We don't know! But we're working on it. We're using molecular genetics to dissect the biosynthetic gene cluster of the TMcin BGC.
Antimicrobial Peptide Mechanism
TMcin kills Gram-positive bacteria by forming large oligomeric pores in bacterial cell membranes. To better take advantage of these pores we're studying how this process works.
People

Sumana Bhowmick - Postdoctoral fellow

Sumana Bhowmick is a postdoctoral fellow and is working to develop new genetic approaches to uncover MRSA physiology and virulence pathways.
Ama Antwi - MOCB

Ama is a graduate student using biochemical and biophysical approaches to understand how TMcin forms pores.
Fauzia Nur - CBSC

Fauzia is graduate student who is interested in understanding how TMcins are produced.
Selected Publications
Seth W. Dickey, Dylan J. Burgin, Ama N. Antwi, Amer Villaruz, Madeline R. Galac, Gordon Y.C. Cheung, Tatiana K. Rostovtseva, Liam J. Worrall, Aleksander C. Lazarski, Elio A. Cino, D. Peter Tieleman, Sergey M. Bezrukov, Natalie C.J. Strynadka, and Michael Otto. Antimicrobial peptide class that forms discrete β-barrel stable pores anchored by transmembrane helices. Nat Commun. 2025 Aug 6;16(1):7231. doi: 10.1038/s41467-025-62604-1. PMID: 40769975; PMCID: PMC12328743.
Seth W. Dickey, Dylan J. Burgin, Steven Huang, David Maguire, and Michael Otto. Two transporters cooperate to secrete amphipathic peptides from the cytoplasmic and membranous milieus. Proceedings of the National Academy of Sciences of the United States of America. 2023 Feb 21;120(8):e2211689120. doi: 10.1073/pnas.2211689120. Epub 2023 Feb 14. PMID: 36787359; PMCID: PMC9974410.
Natalie Zeytuni*, Seth W. Dickey*, Jinhong Hu, H.T. Chou, Liam J. Worrall, J. Andrew N. Alexander, Michael L. Carlson, Michael Nosella, Franck Duong, Z. Yu, Michael Otto, and Natalie C. J. Strynadka, Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins. Sci Adv. 2020;6(40). doi: 10.1126/sciadv.abb8219. PubMed PMID: 32998902; PMCID: PMC7527219. *Authors contributed equally
- Seth W. Dickey, Gordon Y. C. Cheung, and Michael Otto, (2017). Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nature Reviews Drug Discovery, 16(7):457-471. doi: 10.1038/nrd.2017.23. PubMed PMID: 28337021.